Overview¶
This document explains how to run experiments with Seldonian algorithms (SAs) using this library. For a detailed description of what SAs are, see the Seldonian Machine Learning Toolkit homepage.
This library is heavily dependent on the Seldonian Engine library, the core library in the Seldonian Toolkit for running Seldonian algorithms.
Seldonian experiments¶
A Seldonian experiment is a way to evaluate the safety and performance of an SA. It involves running the SA many times with increasing amounts of input data. The way we evaluate safety and performance in this library is with the “Three Plots.”
Three Plots¶
The Three Plots that are generated using this library are:
Performance: the value of some function that indicates performance (e.g., accuracy), evaluated at the solution returned by the SA on a ground truth dataset. Satisfying the behavioral constraints sometimes comes at the cost of reduced performance, and this plot can help you understand that trade-off.
Solution rate: the probability that a solution is returned by the SA. If the behavioral constraints cannot be satisfied given the data provided to the SA, the SA will return No Solution Found.
Failure rate: the probability that a solution is not safe (evaluated on a ground truth dataset) despite the SA returning a solution it determined to be safe, i.e., it passed the safety test.
All three quantities are plotted against the amount of data input to the algorithm on the horizontal axis. Evaluating the performance and failure rate of the algorithm assumes access to ground truth values for performance and safety. For a real-world problem with only a single data set, one typically does not have access to these ground truth quantities. Instead, one can adopt strategies such as bootstrap resampling of their single dataset in order to approximate ground truth.
Figure 1 shows the Three Plots from the GPA prediction problem discussed in the Seldonian Science paper (Figure 3, top panel).
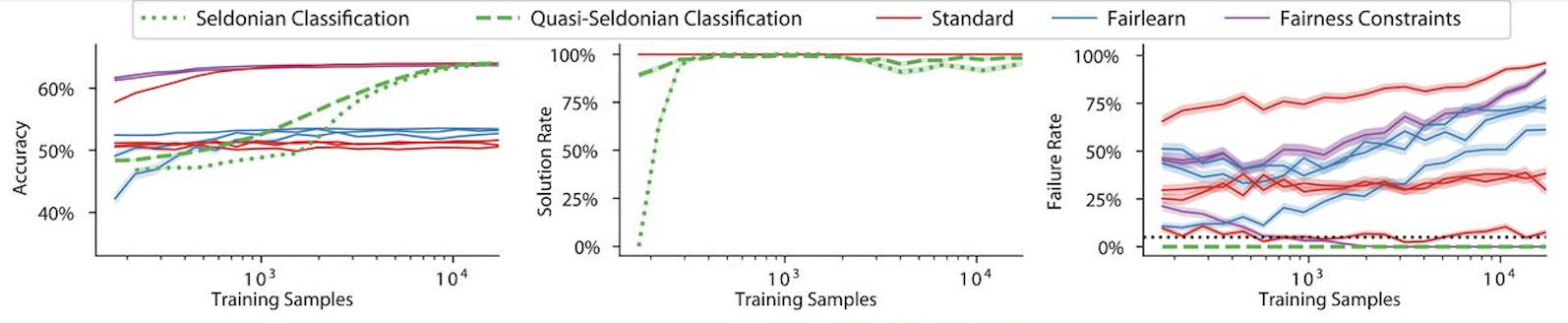
Figure 1: Accuracy (left), solution rate (middle), and failure rate (right) plotted as a function of number of training samples for GPA prediction problem discussed by Thomas et al. (2019). Ground truth was approximated using bootstrap resampling of the original dataset. The fairness constraint in this case is disparate impact. Two Seldonian algorithms, Seldonian Classification (green dotted) and Quasi-Seldonian Classification (green dashed), are compared to several standard ML classification algorithms (red) that do not include the fairness constraint. Also shown are two fairness-aware libraries, Fairlearn (blue) and Fairness Constraints (magenta). In this example, only Seldonian algorithms satisfy the disparate impact criteria (right). The black dotted line in right panel represents the confidence threshold, \({\delta}=0.05\), used in the constraint.¶
Plot generator¶
Depending on the regime of your problem, i.e., supervised learning or reinforcement learning (RL), the object used to produce the Three Plots is either SupervisedPlotGenerator or RLPlotGenerator. While the inputs for both of these classes are described in the API documentation, we will describe their inputs in more detail here.
Regardless of regime, the following inputs are required:
Spec object¶
Often, a Seldonian interface is used to create the Spec object. The Spec object contains everything that is needed to run the SA, such as the original dataset, the parse trees (containing the behavioral constraints), the underlying machine learning model, etc…
n_trials¶
The number of times the SA is run for each amount of data (point on the horizontal axis, see: data_fracs). Used to estimate uncertainties in the quantities in the three plots.
data_fracs¶
A list of fractions of the original dataset size at which to run the SA n_trials times. This list of fractions, multiplied by the number of points in the original dataset, comprises the horizontal axis of each of the three plots. The original dataset is contained within the Spec object.
datagen_method¶
The method for generating data that is used to run the Seldonian algorithm for each trial. For supervised learning, the only currently supported option for this parameter is “resample”. In this case, the original dataset is bootstrap resampled (with replacement) n_trials times to obtain n_trials different datasets of the same dimensions as the original dataset. At each data fraction, frac, in data_fracs, the first frac fraction of points in each of the n_trials datasets is used as input to the SA.
For RL, the only currently supported option for this parameter is “generate_episodes”. In this case, n_trials new datasets are generated with the same number of episodes as the original dataset using the behavior policy. At each data fraction, frac, in data_fracs, the first frac fraction of episodes in each of the n_trials generated datasets is used as input to the SA.
n_workers¶
The number of parallel workers to use when running an experiment, if multiple cores are available on the machine running the experiment. Because each trial is independent of all other trials, Seldonian experiments are embarrassingly parallel programs. If the number of cores on the machine running the experiment is less than n_workers, then the max number of cores available will be used.
perf_eval_fn¶
The function or method used to evaluate the performance of the SA in each trial (plot 1/3). This can be the same as the primary objective specified in the Spec object, but it must be explicitly specified. The only required input to this function is the solution returned by the SA. If NSF is returned for a given trial, then this function will not be evaluated for that trial.
perf_eval_kwargs¶
If the perf_eval_fn has more arguments than the solution, pass them as a dictionary in this parameter.
constraint_eval_fns¶
In order to make plot 3/3 (failure rate) the behavioral constraints are evaluated on a ground truth dataset. If this parameter is left as an empty list (default), the constraints will be evaluated using built-in methods in the parse trees. If instead you have custom functions that you want to use to evaluate the behavioral constraints, pass them as a list in this parameter. The list must be the same length as the number of behavioral constraints.
constraint_eval_kwargs¶
If your constraint_eval_fns have more arguments than the solution returned by the SA, pass them as a dictionary in this parameter.
results_dir¶
The directory in which to save the results of the experiment.
An example API call to make the three plots in the supervised learning regime is:
plot_generator = SupervisedPlotGenerator(
spec=spec,
n_trials=n_trials,
data_fracs=data_fracs,
n_workers=n_workers,
datagen_method='resample',
perf_eval_fn=perf_eval_fn,
constraint_eval_fns=[],
results_dir=results_dir,
perf_eval_kwargs=perf_eval_kwargs,
)
plot_generator.run_seldonian_experiment()
plot_generator.make_plots()
Baselines¶
Often one wants to evaluate the accuracy, solution rate and failure rate of their Seldonian algorithm against that of other machine learning models. In this library, we call all of these other models baselines. We created the BaselineExperiment class to make it easy for developers to easily add their own baseline models. We added several example baselines already, including logistic regression and a random classifier which predicts the positive class with \(p=0.5\) every time. For example, to run the same experiment above including a baseline logistic regression experiment we would simply add one line to the code block above, like:
plot_generator = SupervisedPlotGenerator(
spec=spec,
n_trials=n_trials,
data_fracs=data_fracs,
n_workers=n_workers,
datagen_method='resample',
perf_eval_fn=perf_eval_fn,
constraint_eval_fns=[],
results_dir=results_dir,
perf_eval_kwargs=perf_eval_kwargs,
)
plot_generator.run_seldonian_experiment()
plot_generator.run_baseline_experiment(model_name='logistic_regression')
plot_generator.make_plots()
We use the same plot generator for both the Seldonian model and the baseline model because we want to use the same parameters for both experiments, such as n_trials and data_fracs. That way, they can be compared against each other in the Three Plots.
We also created a .FairlearnExperiment class which implements another type of fairness-aware machine learning model called Fairlearn. Fairlearn experiments are currently only supported for classification problems for a narrow range of behavioral constraints. The documentation for these experiments is currently in progress.
Files generated in an experiment¶
The directory structure inside results_dir will look like this after running a Seldonian experiment via plot_generator.run_seldonian_experiment():
├── qsa_results
│ ├── qsa_results.csv
│ └── trial_data
│ ├── data_frac_0.0010_trial_0.csv
│ ├── data_frac_0.0010_trial_1.csv
│ ├── data_frac_0.0010_trial_2.csv
│ ├── data_frac_0.0010_trial_3.csv
│ ├── data_frac_0.0010_trial_4.csv
│ ├── data_frac_0.0022_trial_0.csv
│ ├── data_frac_0.0022_trial_1.csv
│ ├── data_frac_0.0022_trial_2.csv
│ ├── data_frac_0.0022_trial_3.csv
│ ├── data_frac_0.0022_trial_4.csv
│ ├── data_frac_0.0046_trial_0.csv
│ ├── data_frac_0.0046_trial_1.csv
│ ├── data_frac_0.0046_trial_2.csv
│ ├── data_frac_0.0046_trial_3.csv
│ ├── data_frac_0.0046_trial_4.csv
│ ├── data_frac_0.0050_trial_0.csv
│ ├── data_frac_0.0100_trial_0.csv
│ ├── data_frac_0.0100_trial_1.csv
│ ├── data_frac_0.0100_trial_2.csv
│ ├── data_frac_0.0100_trial_3.csv
│ ├── data_frac_0.0100_trial_4.csv
│ ├── data_frac_0.0215_trial_0.csv
│ ├── data_frac_0.0215_trial_1.csv
│ ├── data_frac_0.0215_trial_2.csv
│ ├── data_frac_0.0215_trial_3.csv
│ ├── data_frac_0.0215_trial_4.csv
│ ├── data_frac_0.0464_trial_0.csv
│ ├── data_frac_0.0464_trial_1.csv
│ ├── data_frac_0.0464_trial_2.csv
│ ├── data_frac_0.0464_trial_3.csv
│ ├── data_frac_0.0464_trial_4.csv
│ ├── data_frac_0.1000_trial_0.csv
│ ├── data_frac_0.1000_trial_1.csv
│ ├── data_frac_0.1000_trial_2.csv
│ ├── data_frac_0.1000_trial_3.csv
│ ├── data_frac_0.1000_trial_4.csv
│ ├── data_frac_0.2154_trial_0.csv
│ ├── data_frac_0.2154_trial_1.csv
│ ├── data_frac_0.2154_trial_2.csv
│ ├── data_frac_0.2154_trial_3.csv
│ ├── data_frac_0.2154_trial_4.csv
│ ├── data_frac_0.4642_trial_0.csv
│ ├── data_frac_0.4642_trial_1.csv
│ ├── data_frac_0.4642_trial_2.csv
│ ├── data_frac_0.4642_trial_3.csv
│ ├── data_frac_0.4642_trial_4.csv
│ ├── data_frac_1.0000_trial_0.csv
│ ├── data_frac_1.0000_trial_1.csv
│ ├── data_frac_1.0000_trial_2.csv
│ ├── data_frac_1.0000_trial_3.csv
│ ├── data_frac_1.0000_trial_4.csv
└── resampled_datasets
├── resampled_data_trial0.pkl
├── resampled_data_trial1.pkl
├── resampled_data_trial2.pkl
├── resampled_data_trial3.pkl
├── resampled_data_trial4.pkl
In this example experiment, n_trials=5 and the default was used for data_fracs, i.e., np.logspace(-3,0,10), which creates a log-spaced array of length 10 starting at \(10^{-3}=0.001\) and ending at \(10^0=1.0\). As a result, there are \(5*10=50\) CSV files created in trial_data/. Each of these CSV files contains the performance, a boolean flag for whether the solution passed the safety test, and a boolean flag for whether the solution failed on the ground truth data set for the given trial at the given data fraction. For example, the contents of the file data_frac_0.6105_trial36.csv are:
data_frac,trial_i,performance,passed_safety,failed
0.6105402296585326,36,0.6746247014792527,True,False
For each baseline experiment run, a new top level folder in results_dir will be created with the name {model_name}_results. For example if a logistic regression baseline was run with the code: plot_generator.run_baseline_experiment(model_name='logistic_regression'), the folder will be called logistic_regression_results and will have the contents:
├── logistic_regression_results
│ ├── logistic_regression_results.csv
│ └── trial_data
│ ├── data_frac_0.0010_trial_0.csv
│ ├── data_frac_0.0010_trial_1.csv
│ ├── data_frac_0.0010_trial_2.csv
│ ├── data_frac_0.0010_trial_3.csv
│ ├── data_frac_0.0010_trial_4.csv
│ ├── data_frac_0.0022_trial_0.csv
│ ├── data_frac_0.0022_trial_1.csv
│ ├── data_frac_0.0022_trial_2.csv
│ ├── data_frac_0.0022_trial_3.csv
│ ├── data_frac_0.0022_trial_4.csv
│ ├── data_frac_0.0046_trial_0.csv
│ ├── data_frac_0.0046_trial_1.csv
│ ├── data_frac_0.0046_trial_2.csv
│ ├── data_frac_0.0046_trial_3.csv
│ ├── data_frac_0.0046_trial_4.csv
│ ├── data_frac_0.0050_trial_0.csv
│ ├── data_frac_0.0100_trial_0.csv
│ ├── data_frac_0.0100_trial_1.csv
│ ├── data_frac_0.0100_trial_2.csv
│ ├── data_frac_0.0100_trial_3.csv
│ ├── data_frac_0.0100_trial_4.csv
│ ├── data_frac_0.0215_trial_0.csv
│ ├── data_frac_0.0215_trial_1.csv
│ ├── data_frac_0.0215_trial_2.csv
│ ├── data_frac_0.0215_trial_3.csv
│ ├── data_frac_0.0215_trial_4.csv
│ ├── data_frac_0.0464_trial_0.csv
│ ├── data_frac_0.0464_trial_1.csv
│ ├── data_frac_0.0464_trial_2.csv
│ ├── data_frac_0.0464_trial_3.csv
│ ├── data_frac_0.0464_trial_4.csv
│ ├── data_frac_0.1000_trial_0.csv
│ ├── data_frac_0.1000_trial_1.csv
│ ├── data_frac_0.1000_trial_2.csv
│ ├── data_frac_0.1000_trial_3.csv
│ ├── data_frac_0.1000_trial_4.csv
│ ├── data_frac_0.2154_trial_0.csv
│ ├── data_frac_0.2154_trial_1.csv
│ ├── data_frac_0.2154_trial_2.csv
│ ├── data_frac_0.2154_trial_3.csv
│ ├── data_frac_0.2154_trial_4.csv
│ ├── data_frac_0.4642_trial_0.csv
│ ├── data_frac_0.4642_trial_1.csv
│ ├── data_frac_0.4642_trial_2.csv
│ ├── data_frac_0.4642_trial_3.csv
│ ├── data_frac_0.4642_trial_4.csv
│ ├── data_frac_1.0000_trial_0.csv
│ ├── data_frac_1.0000_trial_1.csv
│ ├── data_frac_1.0000_trial_2.csv
│ ├── data_frac_1.0000_trial_3.csv
│ ├── data_frac_1.0000_trial_4.csv
When plot_generator.make_plots() is called, it will look for all folders in results_dir ending with _results. Each folder will be considered its own model and will be plotted as a separate curve in the Three Plots figure.
For an end-to-end example use case that makes use of the Seldonian Experiments library, see: Fairness in Automated Loan Approval Systems tutorial.